Contents
close all
clear variables
Dataset specification and parameters
fitType = 2
crop = true;
shouldSaveResults = 1;
format short e
for dataset = 30
warning('off','all')
rmpath(genpath('Pump-Side\'));
rmpath(genpath('Back Propagation of 1550nm\'));
rmpath(genpath('After Beam Reducer\'));
warning('on','all')
if dataset == 1
type = "Pump-side"
z=[75, 240,500,790,1040,1205]'.*1e-3;
dataPath = 'Pump-Side\After first Flip-Mirror'
names = {'75.PNG','240.PNG','500.PNG','790.PNG','1040.PNG','1205.PNG'}
downsamplingFactor = 8
waistGuess = 3
end
if dataset == 2
type = "Pump-side"
z=[150,250 350,450,550,650,750,850,950]'.*1e-3;
dataPath = 'Pump-Side\After M3 3-7-20'
names = {'15.PNG','25.PNG','35.PNG','45.PNG','55.PNG','65.PNG','75.PNG','85.PNG','95.PNG'};
downsamplingFactor = 8
waistGuess = 3
crop = 0
guess = [1e-3,0]
end
if dataset == 3
type = "Back-propagation"
z = [1,2,3,4,6,7,8,9]'.*1e-1;
dataPath = 'Back Propagation of 1550nm\set3'
names = {'10.pgm','20.pgm','30.pgm','40.pgm','60.pgm','70.pgm','80.pgm','90.pgm'};
downsamplingFactor = 8
waistGuess = 30
end
if dataset == 4
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\3-9-20\'
names = {'Gaussian mode.png'};
downsamplingFactor = 1
waistGuess = 1
end
if dataset == 5
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\4-9-20\'
names = {'1.bmp', '2.bmp','3.bmp','4.bmp','5.bmp'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 6
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\10cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 7
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\30cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 8
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\50cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 9
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\55cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 10
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\60cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 11
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\7-9-20\65cmMore'
names = {'1.png', '2.png','3.png','4.png','5.png'};
downsamplingFactor = 1
waistGuess = 15
end
if dataset == 12
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\8-9-20\'
names = {'6c.png'};
downsamplingFactor = 2
waistGuess = 30
end
if dataset == 13
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\11-9-20\'
names = {'31.png','32.png','33.png'};
downsamplingFactor = 1
waistGuess = 30
end
if dataset == 14
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\11-9-20\'
names = {'15.png'};
downsamplingFactor = 1
waistGuess = 10
end
if dataset == 15
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\Collimator f=15.29mm\25-9-20\'
names = {'1.png','2.png'};
downsamplingFactor = 4
waistGuess = 30
end
if dataset == 16
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\Dichroic at 8cm\'
names = {'85mm.png','86mm.png','90mm.png','105mm.png'};
downsamplingFactor = 2
waistGuess = 30
end
if dataset == 17
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\f=15cm lens\Dichroic at 15cm\'
names = {'95mm.png','96mm.png','105mm.png'};
z=[95,96,105]'*1e-3
downsamplingFactor = 2
waistGuess = 50
end
if dataset == 18
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\f=125mm lens\Dichroic at 15cm\'
names = {'65mm.png','70mm.png','75mm.png'};
downsamplingFactor = 2
waistGuess = 30
end
if dataset == 19
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\f=125mm lens\Dichroic at 125mm\'
names = {'70mm.png','75mm.png'};
downsamplingFactor = 1
waistGuess = 30
end
if dataset == 20
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\f=125mm lens\Dichroic at 80mm\'
names = {'75.png','77.png','80.png'};
downsamplingFactor = 1
waistGuess = 30
crop = false;
end
if dataset == 21
type = "Pump-side"
dataPath = 'Pump-Side\At crystal\28-9-20\'
names = {'85mm.png','150mm.png'};
downsamplingFactor = 1
waistGuess = 10
end
if dataset == 22
type = "Back-propagation"
dataPath = 'Back Propagation of 1550nm\At crystal\28-9-20\f=125mm lens\Dichroic at 135mm\'
names = {'70mm.png','75mm.png','80mm.png'};
downsamplingFactor = 1
waistGuess = 30
end
if dataset == 23
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\At crystal\30-9-20\'
names = {'f=75a.png','f=75b.png','f=75c.png'};
downsamplingFactor = 1
waistGuess = 30
end
if dataset == 24
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=6.24mm\6-10-20\old'
names = {'205.png','365.png','620.png','830.png'};
z = [205,365,620,830]'.*1e-3;
downsamplingFactor = 8
waistGuess = 200
guess = [4e-4 0.4]
end
if dataset == 25
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=6.24mm\6-10-20\8'
names = {'65.png','120.png','225.png','335.png','795.png','1035.png'};
z = [65,120,225,335,795,1035]'.*1e-3;
downsamplingFactor = 4
waistGuess = 100
end
if dataset == 26
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=6.24mm\6-10-20\35'
names = {'95.pgm','350.pgm','820.pgm','1030.pgm'};
z = [95,350,820,1030]'.*1e-3;
downsamplingFactor = 8
waistGuess = 100
guess = [4e-4 0.4]
end
if dataset == 27
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\At crystal\6-10-20\'
names = {'a.png','b.png','c.png'};
downsamplingFactor =1
waistGuess = 30
end
if dataset == 28
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\At crystal\8-10-20\'
names = {'8a.png','8b.png','8c.png'};
downsamplingFactor =1
waistGuess = 30
end
if dataset == 29
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\At crystal\8-10-20\diffgain'
names = {'-5db.png','0db.png','5db.png','10db.png','20dbSat.png'};
downsamplingFactor =1
waistGuess = 30
end
if dataset == 30
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=11mm\12-10-20\Set1\'
names = {'150.png','218.png','487.png','614.png','645.png','732.png','753.png','818.png','90.png'};
z= [150,218,487,614,645,732,753,818,90]'*1e-3
downsamplingFactor = 6
waistGuess = 300
end
if dataset == 31
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=11mm\13-10-20\Set2\'
names = {'145.png', '324.png', '485.png', '518.png', '697.png', '780.png'};
z= [145,324,485,518,697,780]'*1e-3
downsamplingFactor = 6
waistGuess = 300
end
if dataset == 32
type = "Back-propagation";
dataPath = 'Back Propagation of 1550nm\Collimator f=11mm\13-10-20\Set3\'
names = {'166.png', '366.png', '498.png', '601.png', '691.png', '780.png'};
z= [166,366,498,601,691,780]'*1e-3
downsamplingFactor = 6
waistGuess = 300
end
end
fitType =
2
dataPath =
'Back Propagation of 1550nm\Collimator f=11mm\12-10-20\Set1\'
z =
1.5000e-01
2.1800e-01
4.8700e-01
6.1400e-01
6.4500e-01
7.3200e-01
7.5300e-01
8.1800e-01
9.0000e-02
downsamplingFactor =
6
waistGuess =
300
Publish or execute only
shouldPublish = 1
firstPublish = load("firstPublish.mat");
firstPublish = firstPublish.firstPublish;
publishPath = strcat('C:\Users\franc\Box\Dottorato\Projects\SECRET\BeamWidth\Pusblished html\',strrep(dataPath,'\','_'));
if (shouldPublish && firstPublish)
firstPublish = 0;
save("firstPublish.mat","firstPublish")
publish('GaussianBeamFit.m','outputDir',publishPath);
firstPublish = 1;
save("firstPublish.mat","firstPublish");
web(strcat(publishPath,"\GaussianBeamFit.html"));
close all;
else
Sort captures by distance if provided
if exist('z','var')
[z,ordered_ind] = sort(z);
names = names(ordered_ind);
end
Image Loading
addpath(dataPath);
if (type == "Back-propagation")
lambda = 1550e-9
pixel_size = 3.75 *10^(-6)
elseif(type == "Pump-side")
lambda = 775e-9
pixel_size = 5.3e-6
end
n= length(names)
images = cell(n,1);
for i = 1:n
rawData1 = importdata(names{i});
[~,name] = fileparts(names{i});
newData1.(matlab.lang.makeValidName(name)) = rawData1;
vars = fieldnames(newData1);
for j = 1:length(vars)
assignin('base', vars{j}, newData1.(vars{j}));
images{i}= sum(double(eval(vars{j})),3);
end
end
results1 = cell(n);
results2 = cell(n,1);
lambda =
1.5500e-06
pixel_size =
3.7500e-06
n =
9
Show pictures
if 1
for i= 1:length(images)
figure
image(images{i});
title(sprintf('Picture n° %i, dataset n°%d', [i,dataset] ));
end
end

Pre-process pictures
for i = 1:n
img = images{i};
[old_l,old_m] = size(img);
img = imresize(img, 1/downsamplingFactor,'bicubic');
[l,m] = size(img);
downsamplingFactor = sqrt((old_m*old_l)/(l*m));
imgSmoothed = smoothdata(img,'movmedian',20);
[colMax, row] = max(imgSmoothed);
[imgMax, col] = max(colMax);
row = row(col);
if (crop)
width = round( waistGuess / downsamplingFactor) * 2 * 4;
height = round(waistGuess / downsamplingFactor) * 2 * 4;
images{i} = img(1+subplus(row-width/2-1):min(row+width/2,l),1+subplus(col-height/2-1):min(col+height/2,m));
else
images{i} = img;
end
images{i} = images{i}/max(max(images{i}))*100;
end
clear imgMax imgMean colMax row col
Show pre-processed pictures
for i= 1:length(images)
images{i} = double(images{i});
figure
image(images{i})
title(sprintf('Pre-processed picture n° %i, dataset n°%d', [i,dataset] ));
end

Fit Gaussian bell
rsquare_bells = zeros(1,n);
for i= 1:length(images)
clear opts
disp("Fitting image n°" + int2str(i))
img = images{i};
sz = size(img);
x_ax = 1:sz(2);
y_ax = 1:sz(1);
[x,y] = meshgrid(1:sz(2),1:sz(1));
if fitType == 1
opts.tilted= true;
results1{i} = autoGaussianSurf(x,y,img, opts);
end
if fitType ==2
opts = fitoptions( 'Method', 'NonlinearLeastSquares', 'Robust', 'LAR');
opts.Display = 'Off';
opts.StartPoint = [1 100 waistGuess/downsamplingFactor sz(1)/2 sz(2)/2];
[bellFit, bellGof] = gaussian2DFit(y,x,img,opts);
rsquare_bells(i)= bellGof.rsquare;
results2{i} = bellFit;
clear fit
end
end
Fitting image n°1
Fitting image n°2
Fitting image n°3
Fitting image n°4
Fitting image n°5
Fitting image n°6
Fitting image n°7
Fitting image n°8
Fitting image n°9
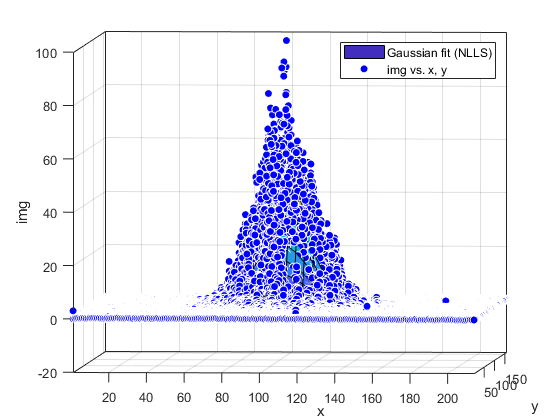
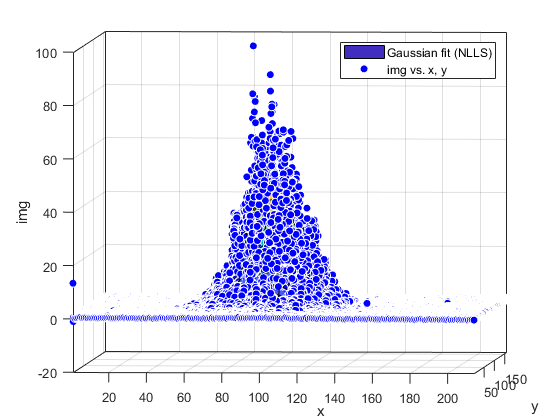
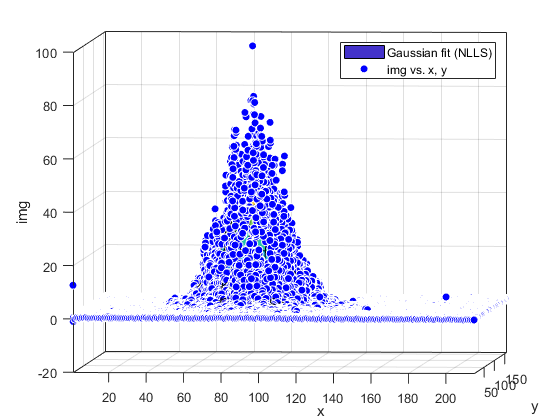
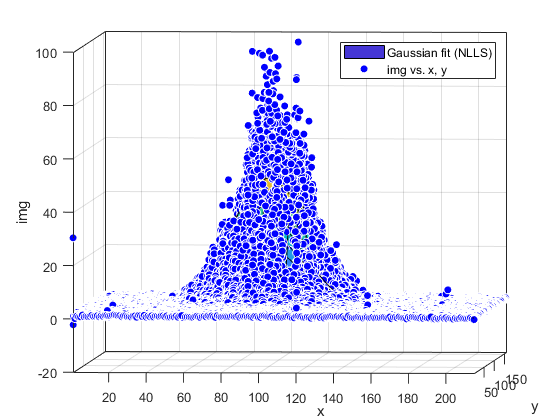

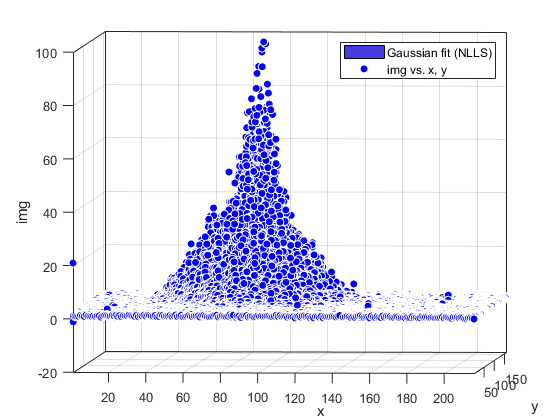
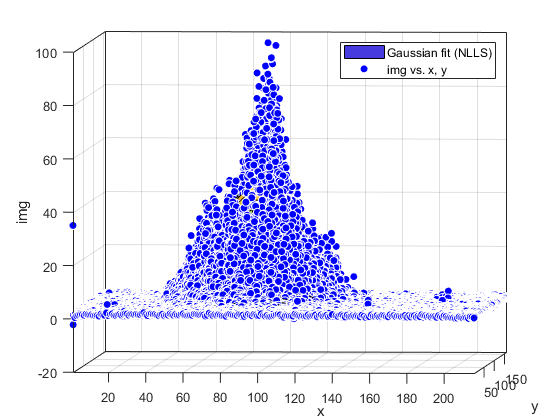

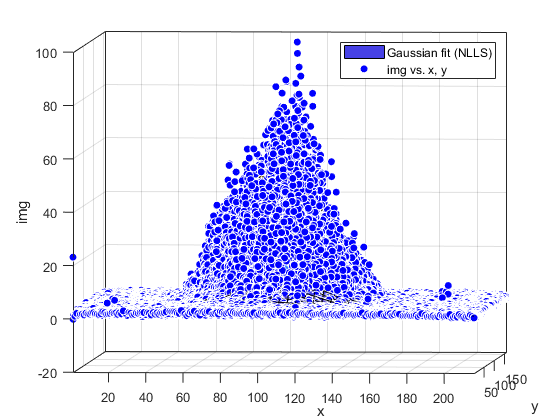
Compute and show residuals
for i= 1:length(images)
sz = size(images{i});
[x,y] = meshgrid(1:sz(2),1:sz(1));
if fitType == 1
r = results1{i};
if (opts.tilted == false)
r.angle = 0;
end
xip = (x-r.x0)*cos(r.angle) + (y-r.y0)*sin(r.angle);
yip =-(x-r.x0)*sin(r.angle) + (y-r.y0)*cos(r.angle);
zfit = r.a*exp(-(xip.^2/2/r.sigmax^2 + ...
yip.^2/2/r.sigmay^2)) + r.b;
end
if fitType == 2
bellFit = results2{i};
zfit= abs(bellFit.A)*exp(-2*((x-abs(bellFit.x0)).^2+(y-abs(bellFit.y0)).^2)/(bellFit.W^2))+bellFit.B;
clear fit
end
if false
figure
image(zfit./max(max(zfit)).*100)
title(sprintf('Fit n° %i, dataset n°%d', [i,dataset] ));
end
figure
image(abs(images{i}-zfit))
title(sprintf('Residuals n° %i, dataset n°%d', [i,dataset] ));
end
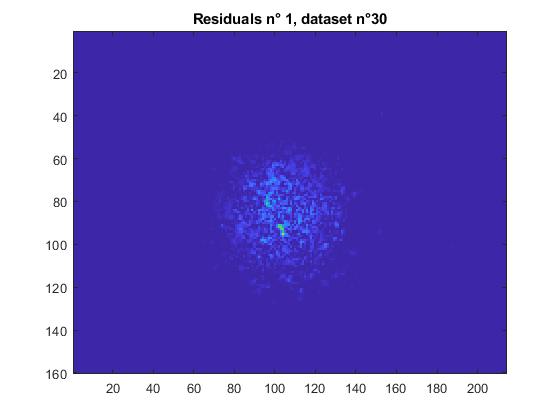


Half diameter computation
variance = zeros(length(images),1);
for i= 1:n
if fitType == 1
variance(i) = results1{i}.sigmax * results1{i}.sigmay;
end
if fitType == 2
variance(i) = results2{i}.W.^2 ./4;
end
end
allW=(4*variance).^(1/2)*pixel_size*downsamplingFactor;
if not(exist('z','var'))
figure1 = figure;
axes1 = axes('Parent',figure1);
plot(allW,'*')
ylabel 'W [m]'
title(sprintf('1/e^2 radius fit of gaussian profiles, dataset n°%d',dataset))
set(axes1,'XTick',1:1:length(allW));
end
Select only images which were fitted correctly
min_rsquare=0.9;
indices = 1:length(images);
chosenIndices = indices;
W=allW(chosenIndices);
Gaussian beam and waist fit
if exist('z','var')
z_selected = z(chosenIndices);
if not(exist('guess','var'))
[smallestW,ind]= min(W)
guess= [smallestW,z_selected(ind)]
end
LAMBDA = num2str(lambda);
modelsTypes = ["ideal gaussian beam model", "imperfect gaussian gaussian beam model (with M^2)"];
for use_real = [0,1]
disp(strcat("Fitting ", modelsTypes(use_real+1)))
if use_real
model= strcat('w_0*M*sqrt( 1+ ( (z-z_i)./( pi*w_0.^2./', LAMBDA,') ).^2 )')
if exist('guess','var')
guess = [1 guess]
end
else
model = strcat('w_0*sqrt( 1+ ( (z-z_i)./( pi*w_0.^2./', LAMBDA,') ).^2 )')
end
ft = fittype( model, 'independent', 'z', 'dependent', 'y' );
opts = fitoptions( 'Method', 'NonlinearLeastSquares');
opts.DiffMaxChange = 1;
opts.DiffMinChange = 1e-09;
opts.Display = 'Off';
opts.MaxIter = 4000;
opts.StartPoint = guess;
[wfit, wgof] = fit( z_selected, W, ft, opts );
rsquare_beam= wgof.rsquare
W_0 = wfit.w_0;
z_i = wfit.z_i;
z_R = pi*(wfit.w_0).^2./lambda ;
theta_0_deg = lambda./pi./wfit.w_0 *(180/pi);
if use_real
M = wfit.M;
theta_0_deg= theta_0_deg * M;
end
beamFigure = figure( 'Name', 'Gaussian beam fit' );
hold on
zstart = -1.5*max([z_R,abs(z_selected(1)-z_i),abs(z_selected(end)-z_i)])+z_i;
zstop = -zstart+2*z_i;
range= zstart:(zstop-zstart)/100:zstop;
plot(z_selected,W,'b*')
plot( range, wfit(range),'r');
plot(range,abs(range-wfit.z_i)*sind(theta_0_deg))
plot(z_selected, wfit(z_selected),'+r')
xlim([zstart,zstop])
vline(wfit.z_i,'k--','waist')
vline(z_i-z_R,'k--','-z_R')
vline(z_i+z_R,'k--','+z_R')
legend('Data', 'NLLS fit','Linear asymptote', 'Location', 'NorthWest' );
xlabel 'z'' [m]'
ylabel '1/e^2 radius [m]'
if use_real
title(sprintf('Non-ideal gaussian beam fit, dataset n°%d',dataset))
else
title(sprintf('Ideal gaussian beam fit, dataset n°%d',dataset))
end
grid on
set(beamFigure, 'HandleVisibility', 'off');
disp('Results in meters:')
W_0
z_i
z_R
theta_0_deg
if use_real
M2 = M^2
W_real = W_0 * M
end
end
smallestW =
6.9498e-04
ind =
1
guess =
6.9498e-04 9.0000e-02
Fitting ideal gaussian beam model
model =
'w_0*sqrt( 1+ ( (z-z_i)./( pi*w_0.^2./1.55e-06) ).^2 )'
rsquare_beam =
8.7356e-01
Results in meters:
W_0 =
6.9682e-04
z_i =
-3.3928e-01
z_R =
9.8414e-01
theta_0_deg =
4.0568e-02
Fitting imperfect gaussian gaussian beam model (with M^2)
model =
'w_0*M*sqrt( 1+ ( (z-z_i)./( pi*w_0.^2./1.55e-06) ).^2 )'
guess =
1.0000e+00 6.9498e-04 9.0000e-02
rsquare_beam =
8.5535e-01
Results in meters:
W_0 =
6.8263e-04
z_i =
-3.0894e-01
z_R =
9.4448e-01
theta_0_deg =
4.2356e-02
M2 =
1.0461e+00
W_real =
6.9821e-04
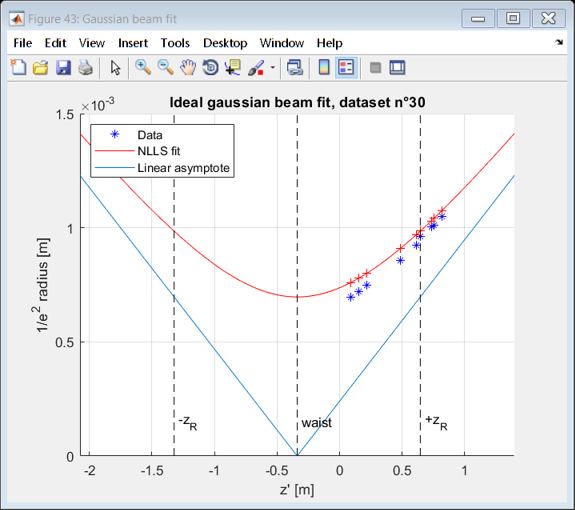
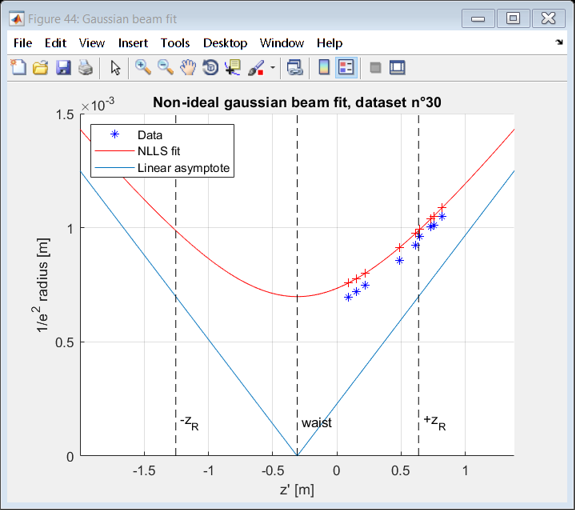
Linear fit
linearFit = false;
if linearFit
pol_fit = fit(z,W,'poly1')
figure
hold on
plot(z,W,'+')
plot(z,pol_fit(z),'-')
xlabel('z'' [m]')
ylabel('W [m]')
title('Polynomial fit')
legend('data','fit')
grid on
theta_lin = atan(pol_fit.p1)
W_0_lin = lambda./pi/theta_lin
z_R_lin = pi*(W_0_lin).^2./lambda
theta_0_lin_deg = lambda./pi./W_0_lin *(180/pi)
end
end
Other expressions (for gaussian beams)
W_chosen = W
STD = W_chosen./2;
FWHM = W_chosen*sqrt(2*log(2));
e2_diam = sqrt(2)*FWHM ./ sqrt(log(2));
W_chosen =
6.9498e-04
7.1862e-04
7.4658e-04
8.5855e-04
9.2301e-04
9.6149e-04
1.0030e-03
1.0094e-03
1.0489e-03
Goodness of fits
rsquare_bells
if exist('z','var')
rsquare_beam
if use_real
M2
end
end
rsquare_bells =
Columns 1 through 6
9.9956e-01 9.9961e-01 9.9960e-01 9.9856e-01 9.9741e-01 9.9682e-01
Columns 7 through 9
9.9460e-01 9.9745e-01 9.9373e-01
rsquare_beam =
8.5535e-01
M2 =
1.0461e+00
Saving results
if shouldSaveResults
outputPath = strcat('Results\',strrep(dataPath,'\','_'))
save(outputPath,'W_chosen','W_0','z_R','z_i','theta_0_deg','results2');
end
outputPath =
'Results\Back Propagation of 1550nm_Collimator f=11mm_12-10-20_Set1_'
end
shouldPublish =
1





































